Bacterial identification
High-resolution bacterial characterisation is essential in microbiology, particularly for disease diagnosis, where rapid and precise identification is a high priority. Gene-based methods have become increasingly important in bacterial classification, complementing and to an extent replacing more traditional phenotypic methods.
However, until now, there has been no single system which works for all bacteria.
Universal gene-based identification
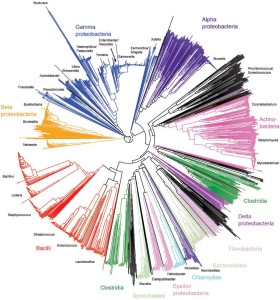
Scientists at the University of Oxford have developed a universal identification scheme based on ribosomal multilocus sequence typing (rMLST).
This represents the first genotypic scheme that can provide both broad and accurate characterisation of bacteria at all phylogenetic levels.
The system works through the identification and analysis of allelic variation within the ribosomal protein subunit (rps) genes, which are universal yet record a wide range of evolutionary diversity, to effect rapid and highly accurate phylogenetic identification.
A web-accessible and expandable database comprising genome data from more than 2000 bacterial isolates has been generated. The variation of 53 rps genes is catalogued in this database, providing a means of defining the precise phylogenetic position of any bacterial sequence at the domain, phylum, class, order, family, genus, species and strain levels.
Benefits of the Oxford system
-
reliable identification
-
high resolution
-
results at the push of a button
-
one system for all bacteria
Applications
The data generated for the rMLST scheme could be used in combination with next-generation sequencing to enable the rapid identification of bacterial isolates at the push of a button. Equally the database enables the development of PCR-based, species or strain-specific diagnostic tests.
Applications include:
-
population studies
-
epidemiological investigations
-
diagnostic tests
about this technology

